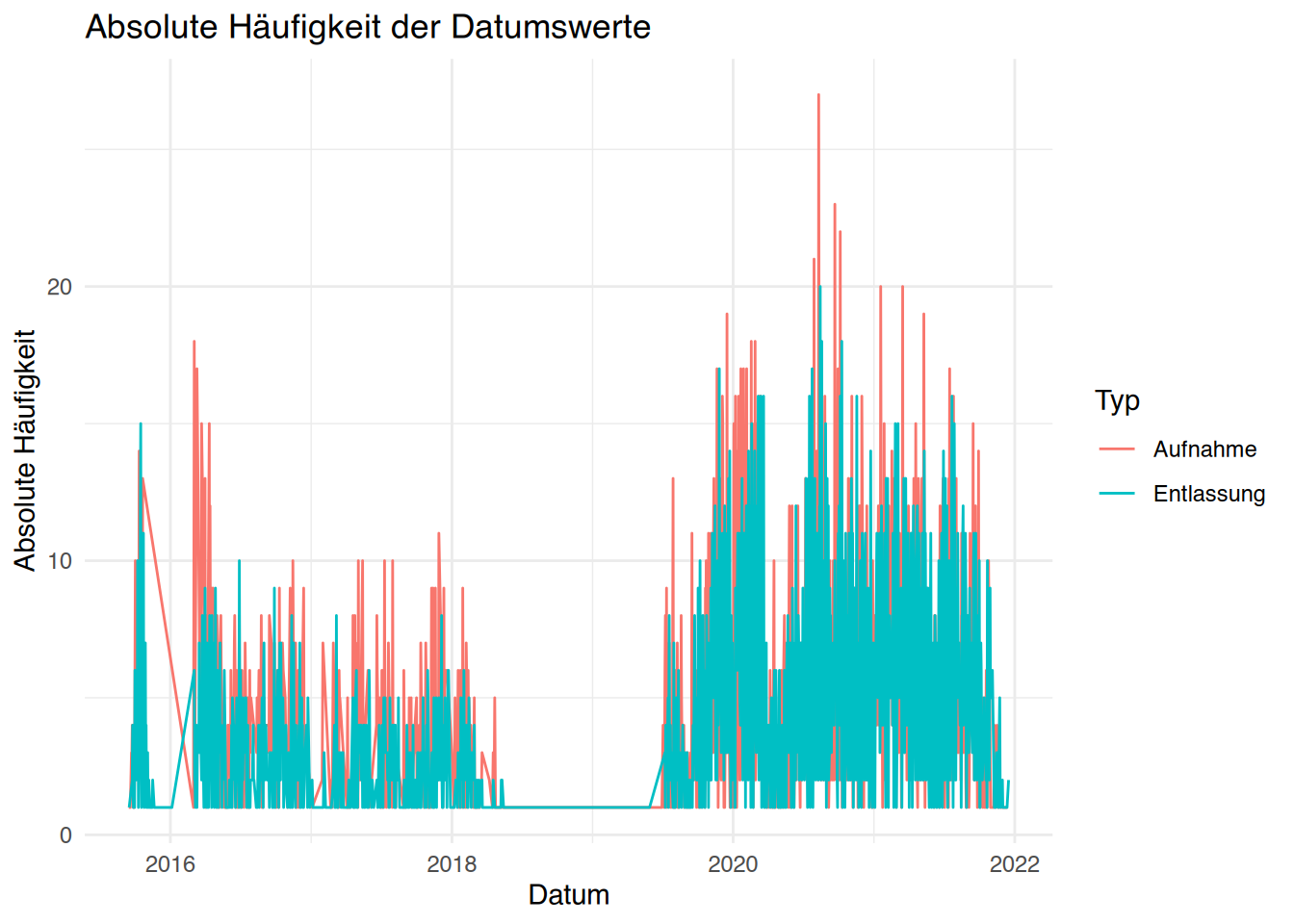
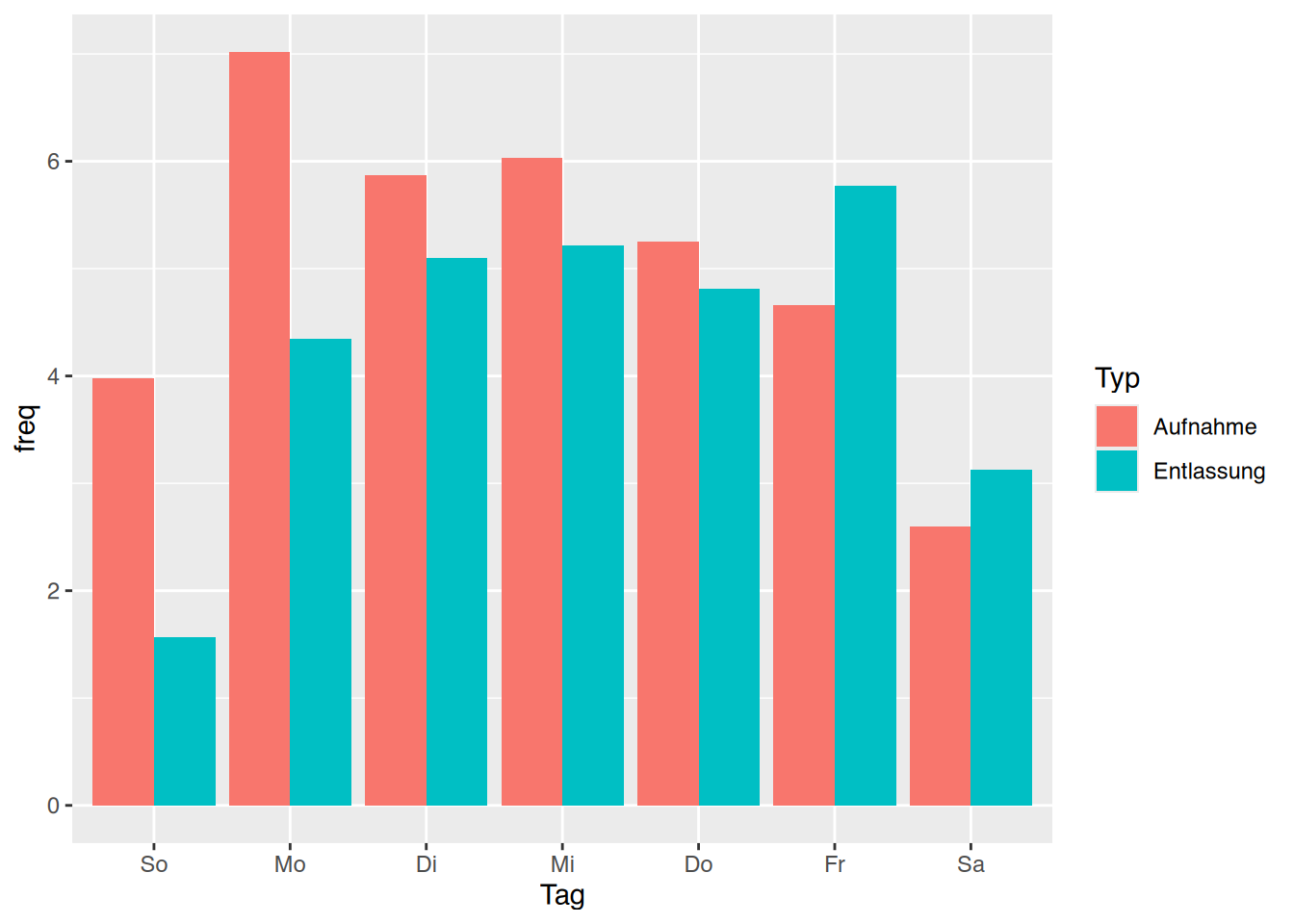
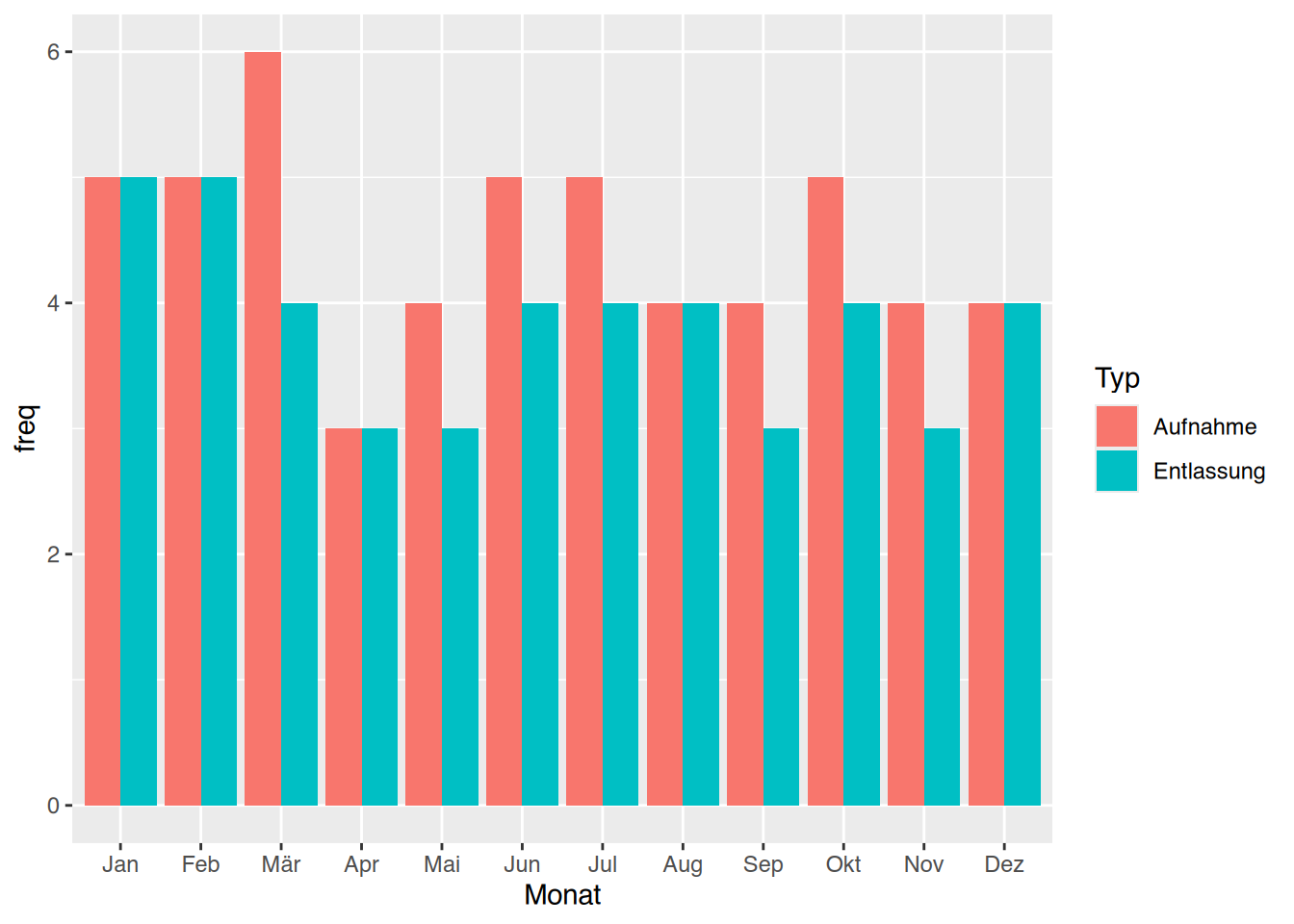
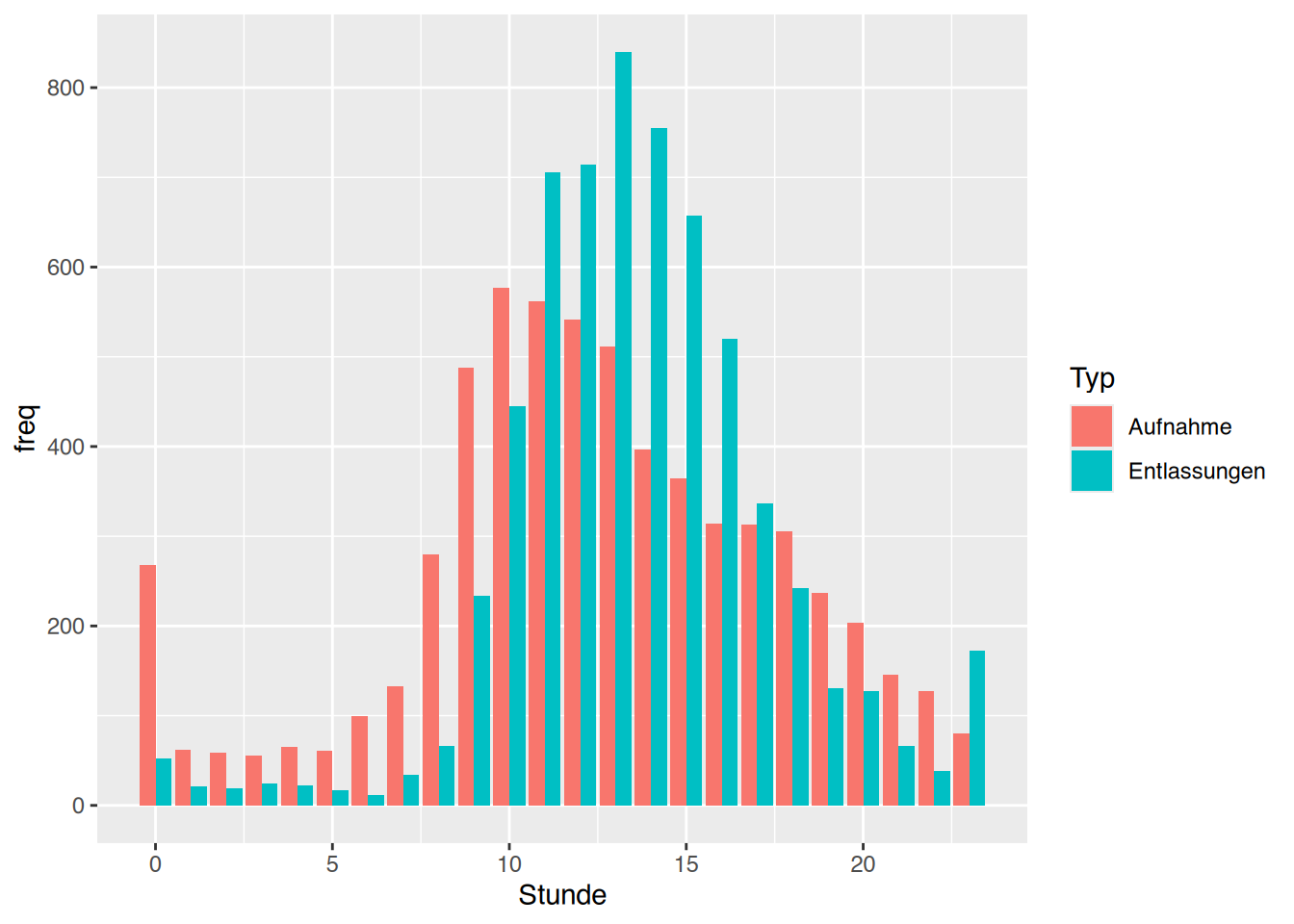
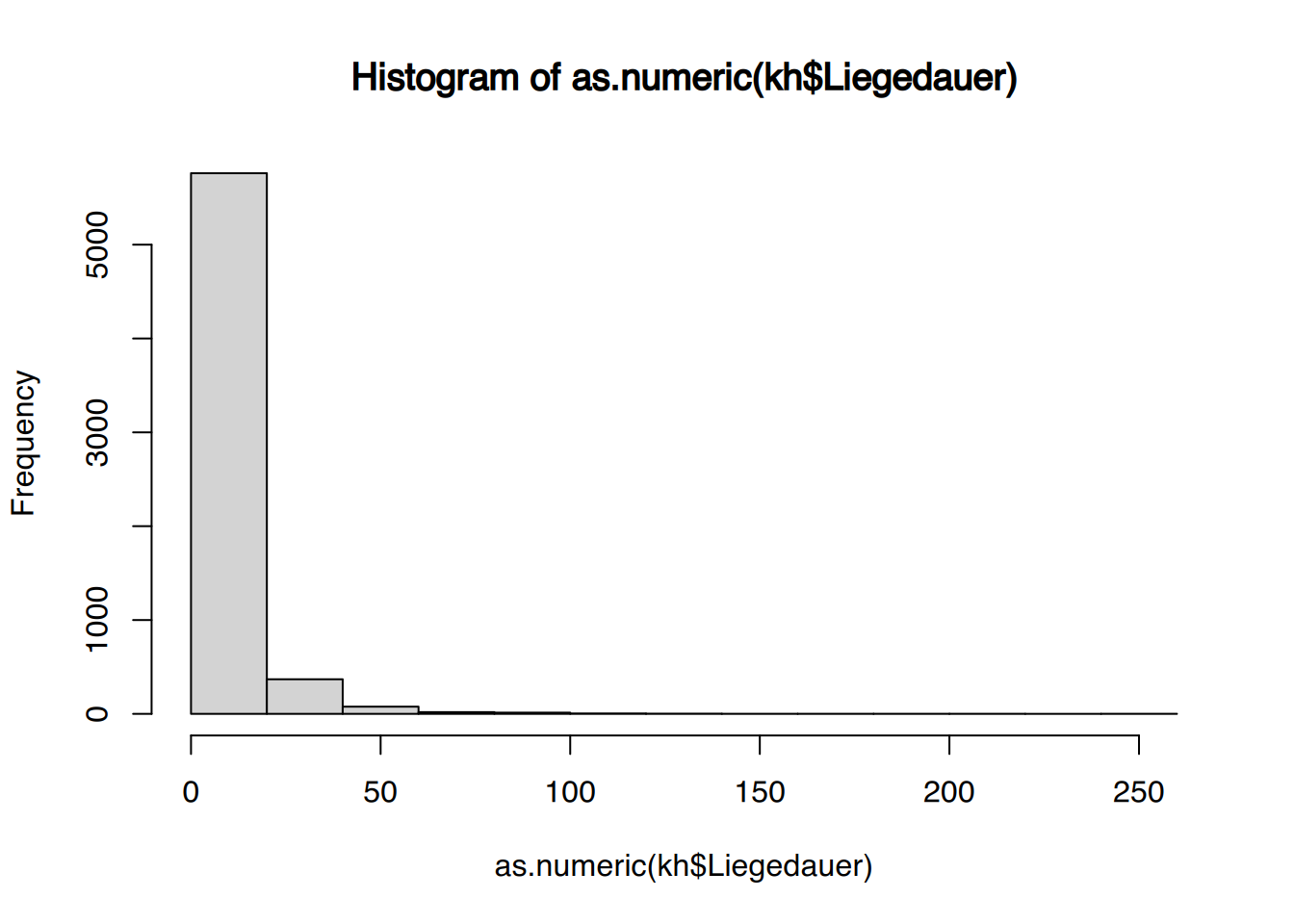
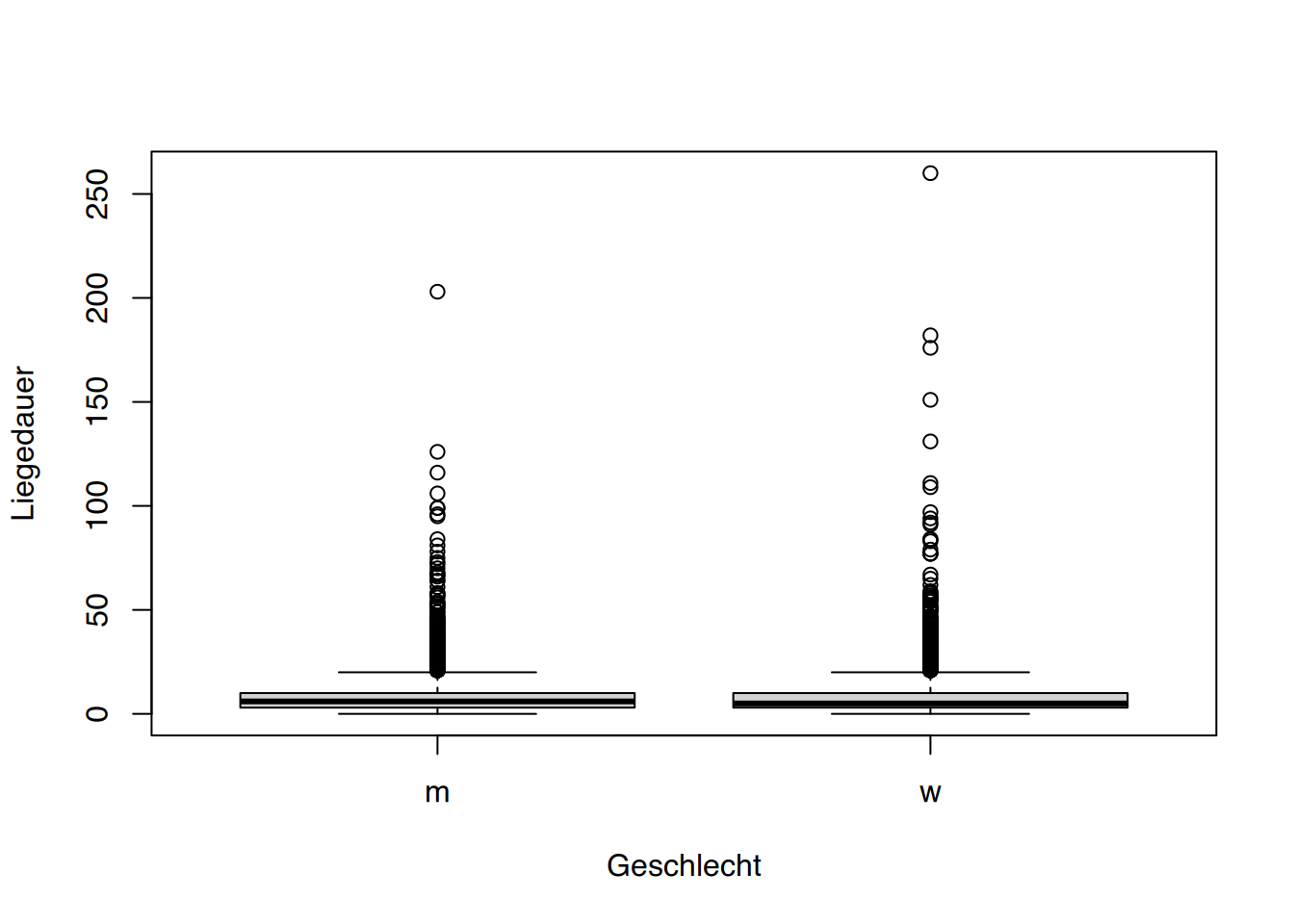
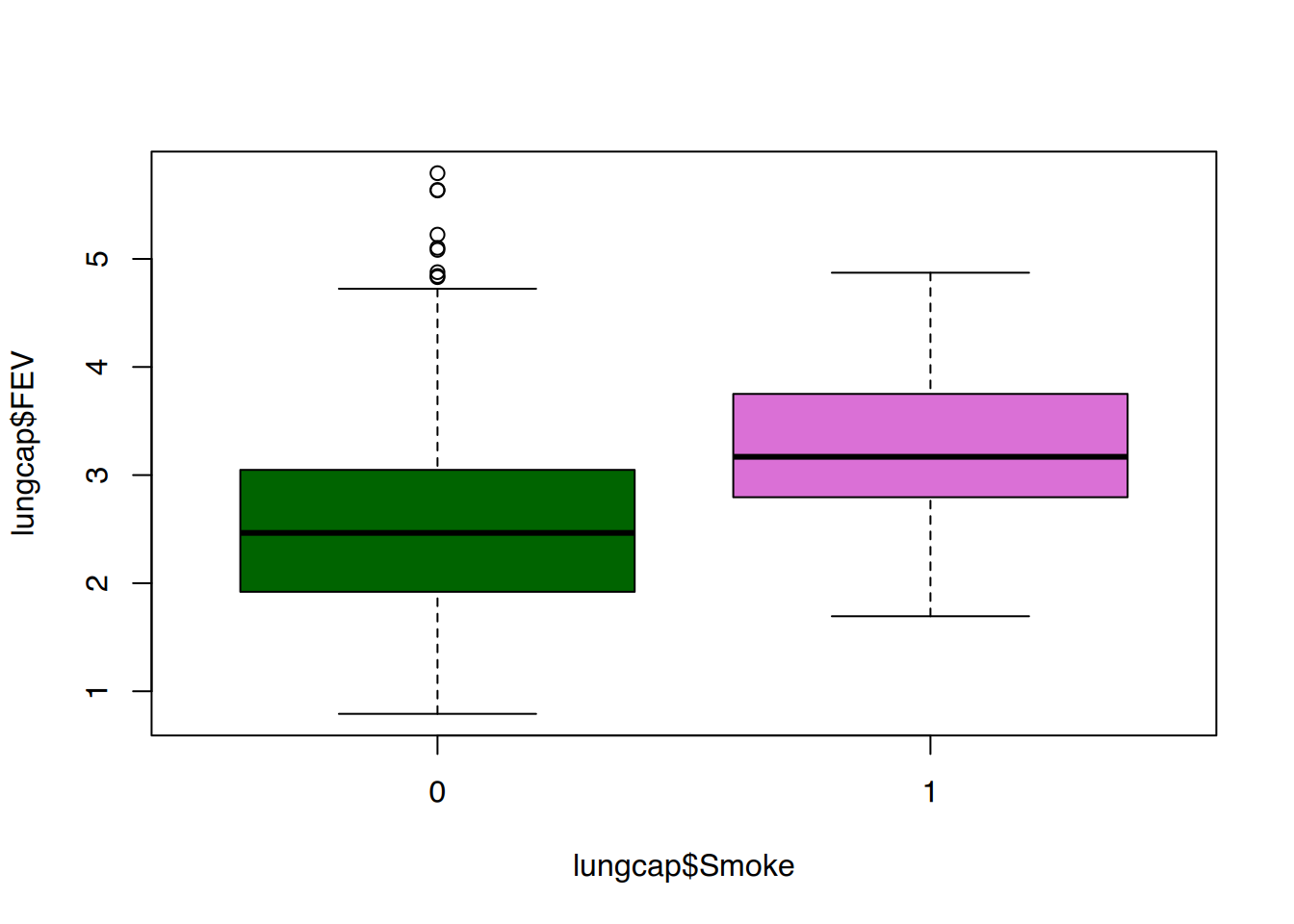
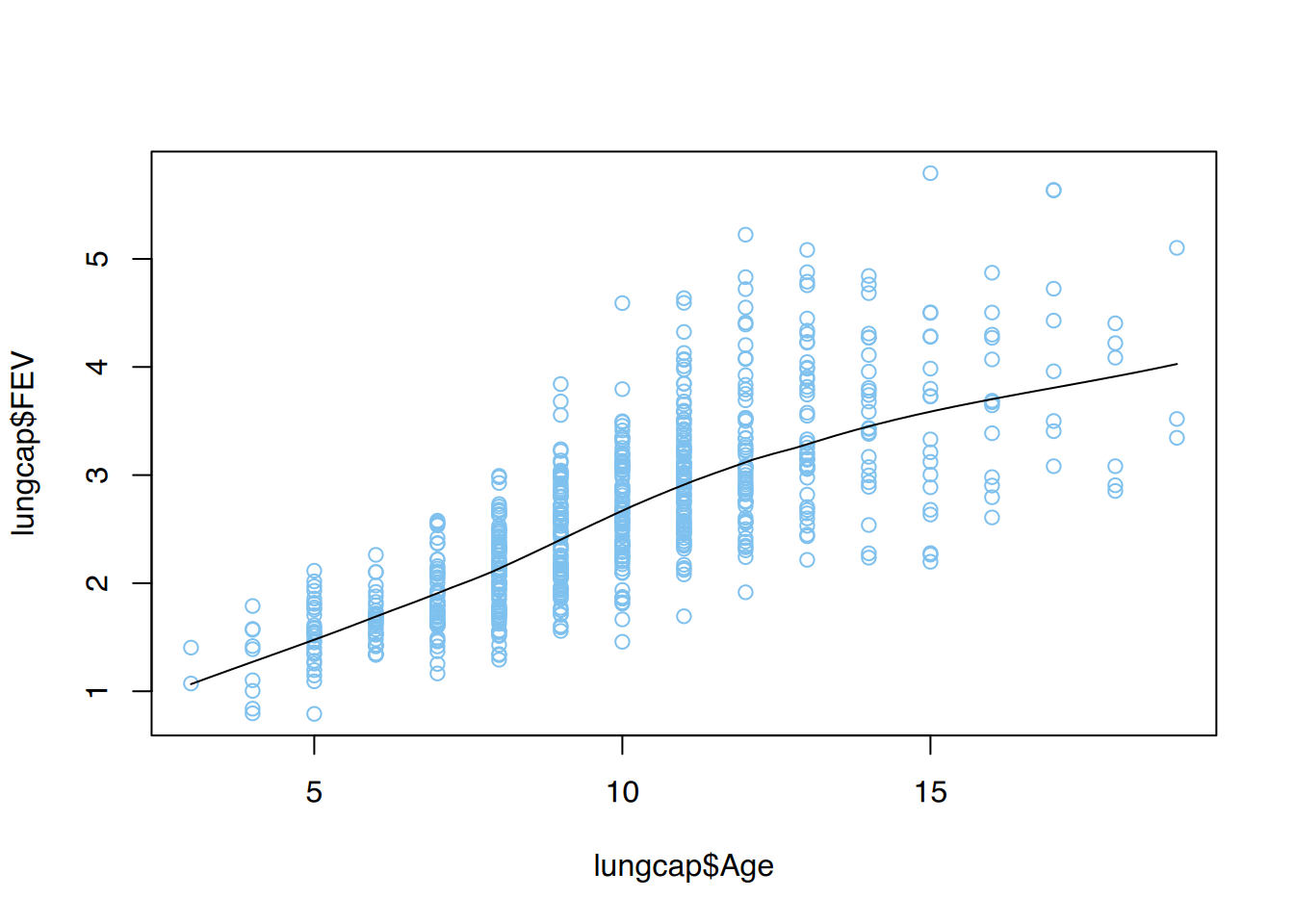
tibble [1,207 × 9] (S3: tbl_df/tbl/data.frame)
$ id : num [1:1207] 1 2 3 4 5 6 7 8 9 10 ...
..- attr(*, "label")= chr "ID"
..- attr(*, "format.spss")= chr "F8.0"
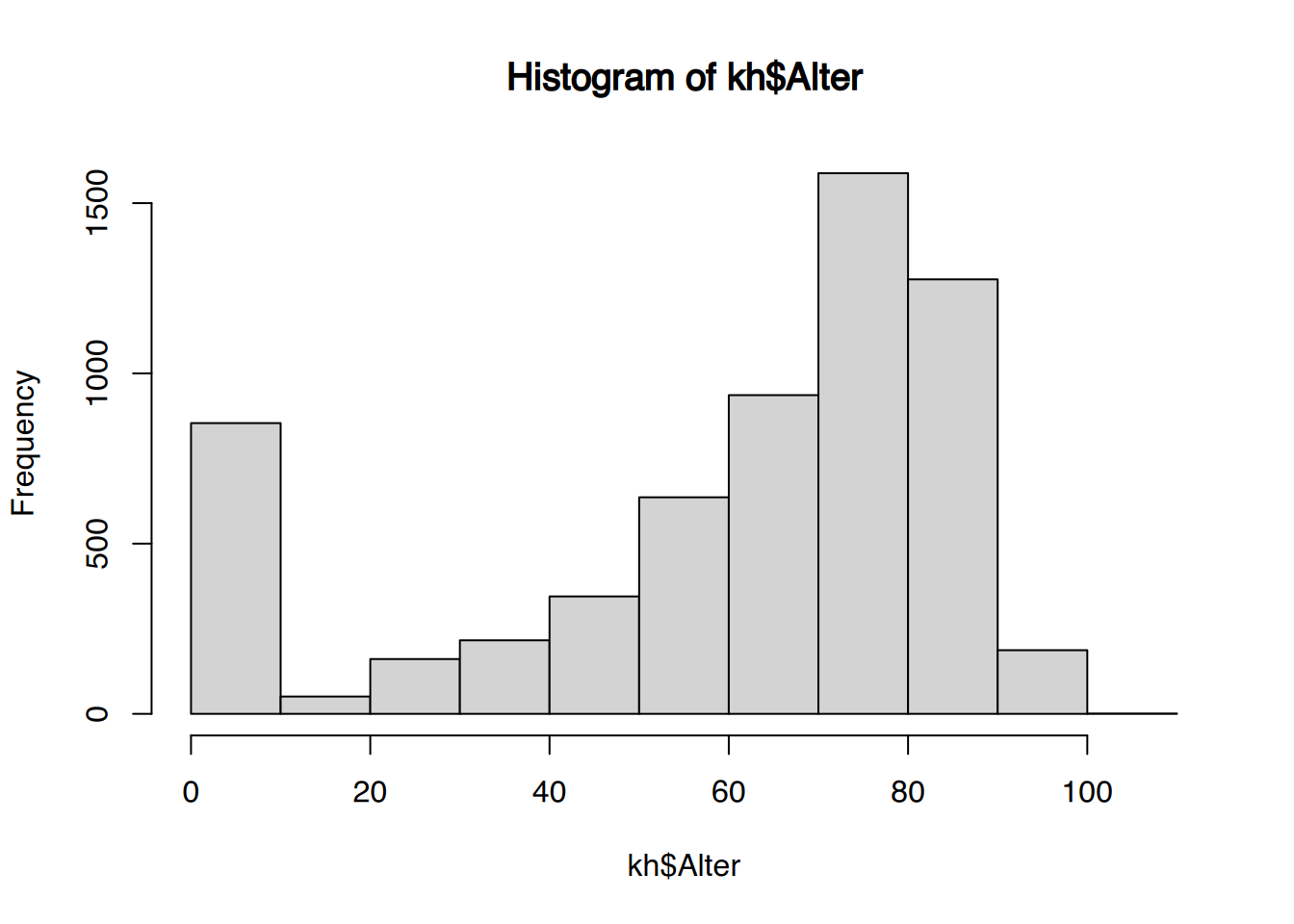
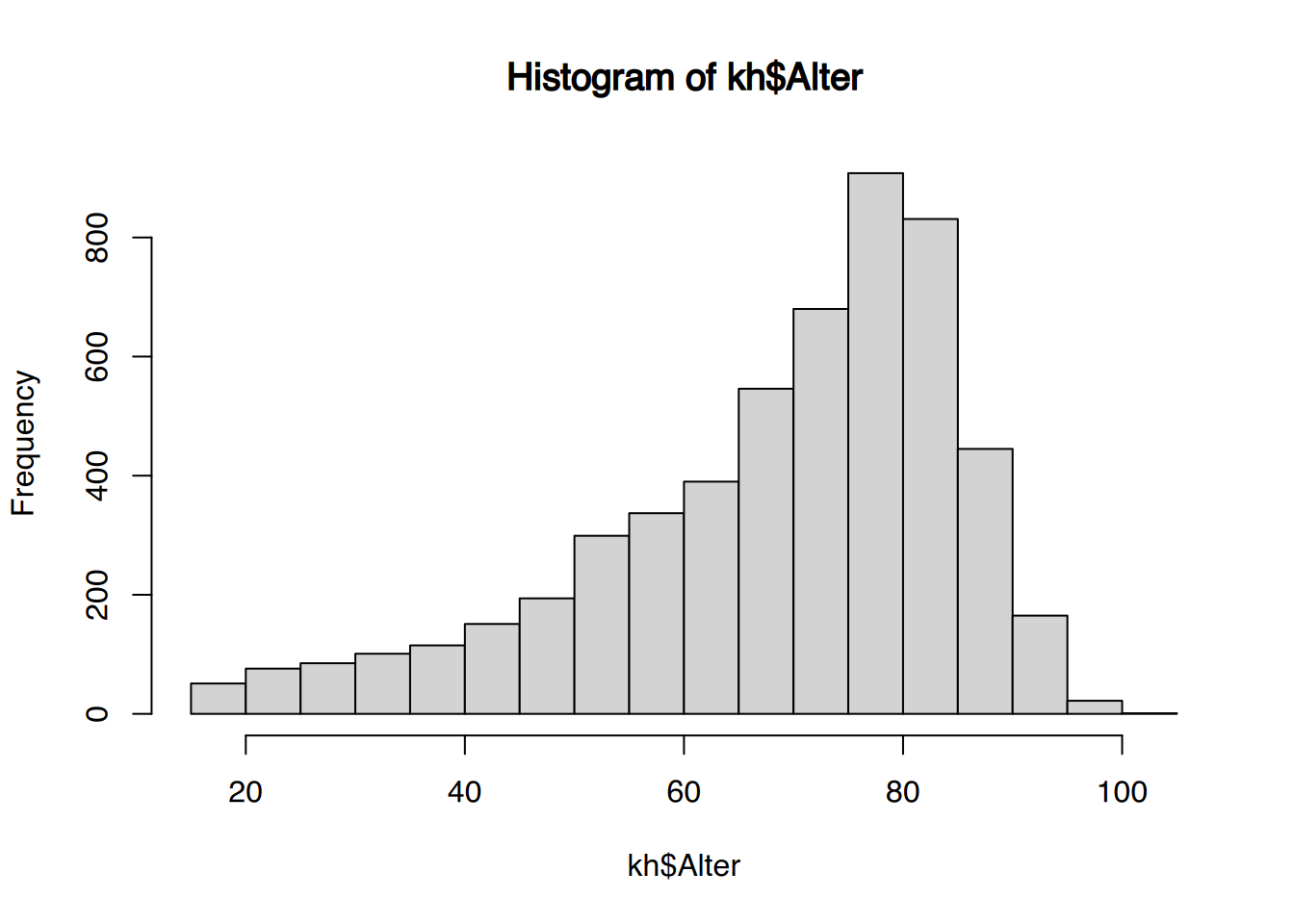
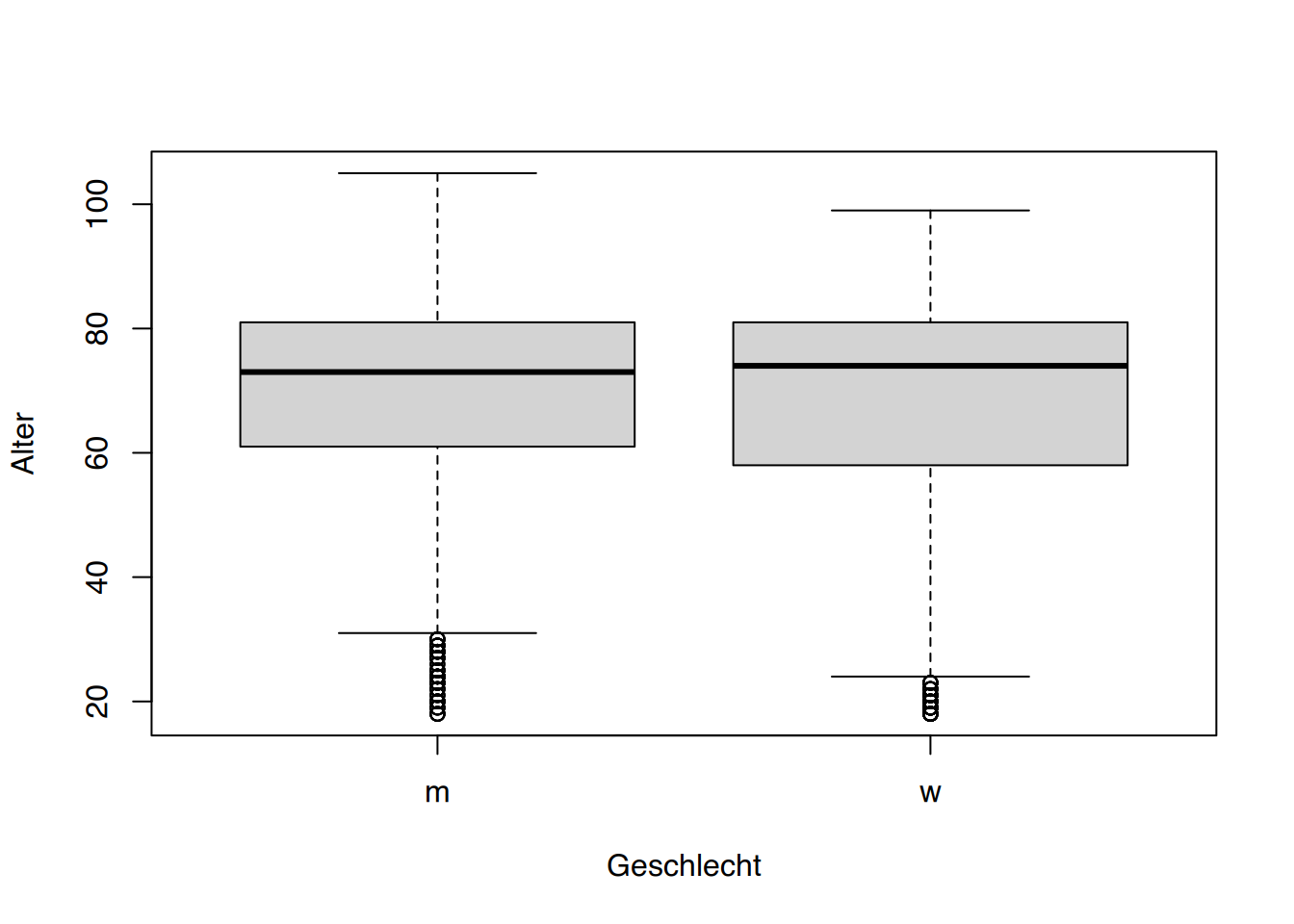
$ age : num [1:1207] 60 79 82 66 52 58 50 83 46 54 ...
..- attr(*, "label")= chr "Alter [Jahre]"
..- attr(*, "format.spss")= chr "F8.0"
$ pathsize: num [1:1207] NA NA NA NA NA NA NA NA NA NA ...
..- attr(*, "label")= chr "Göße des pathologischen Tumors [cm]"
..- attr(*, "format.spss")= chr "F8.2"
$ lnpos : num [1:1207] 0 0 0 0 0 0 0 0 17 6 ...
..- attr(*, "label")= chr "Positive Lymphknoten [Anzahl]"
..- attr(*, "format.spss")= chr "F8.0"
$ histgrad: dbl+lbl [1:1207] 3, 4, 2, 2, 3, 4, 2, 3, 4, 2, 4, 3, 4, 4, 1, 1, 1, 2,...
..@ label : chr "Histologischer Grad"
..@ format.spss: chr "F8.0"
..@ labels : Named num 4
.. ..- attr(*, "names")= chr "Unknown"
$ er : dbl+lbl [1:1207] 0, 2, 2, 1, 2, 2, 1, 0, 2, 1, 2, 2, 2, 2, 1, 2, 2, 2,...
..@ label : chr "Östrogen-Rezeptor-Status"
..@ format.spss: chr "F6.0"
..@ labels : Named num [1:3] 0 1 2
.. ..- attr(*, "names")= chr [1:3] "Negativ" "Positiv" "Unbekannt"
$ pr : dbl+lbl [1:1207] 0, 2, 2, 1, 2, 2, 0, 0, 2, 1, 2, 2, 2, 2, 1, 2, 2, 2,...
..@ label : chr "Progesteron-Rezeptor-Status"
..@ format.spss: chr "F6.0"
..@ labels : Named num [1:3] 0 1 2
.. ..- attr(*, "names")= chr [1:3] "Negativ" "Positiv" "Unbekannt"
$ status : dbl+lbl [1:1207] 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0, 0,...
..@ label : chr "Status"
..@ format.spss: chr "F8.0"
..@ labels : Named num [1:2] 0 1
.. ..- attr(*, "names")= chr [1:2] "Zensiert" "Verstorben"
$ time : num [1:1207] 9.47 8.6 19.33 16.33 8.5 ...
..- attr(*, "label")= chr "Zeit [Monate]"
..- attr(*, "format.spss")= chr "F8.2"